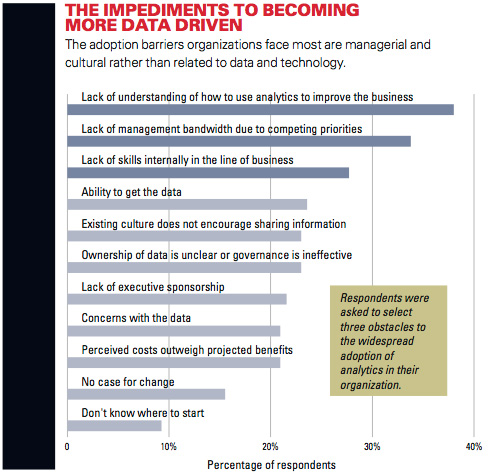
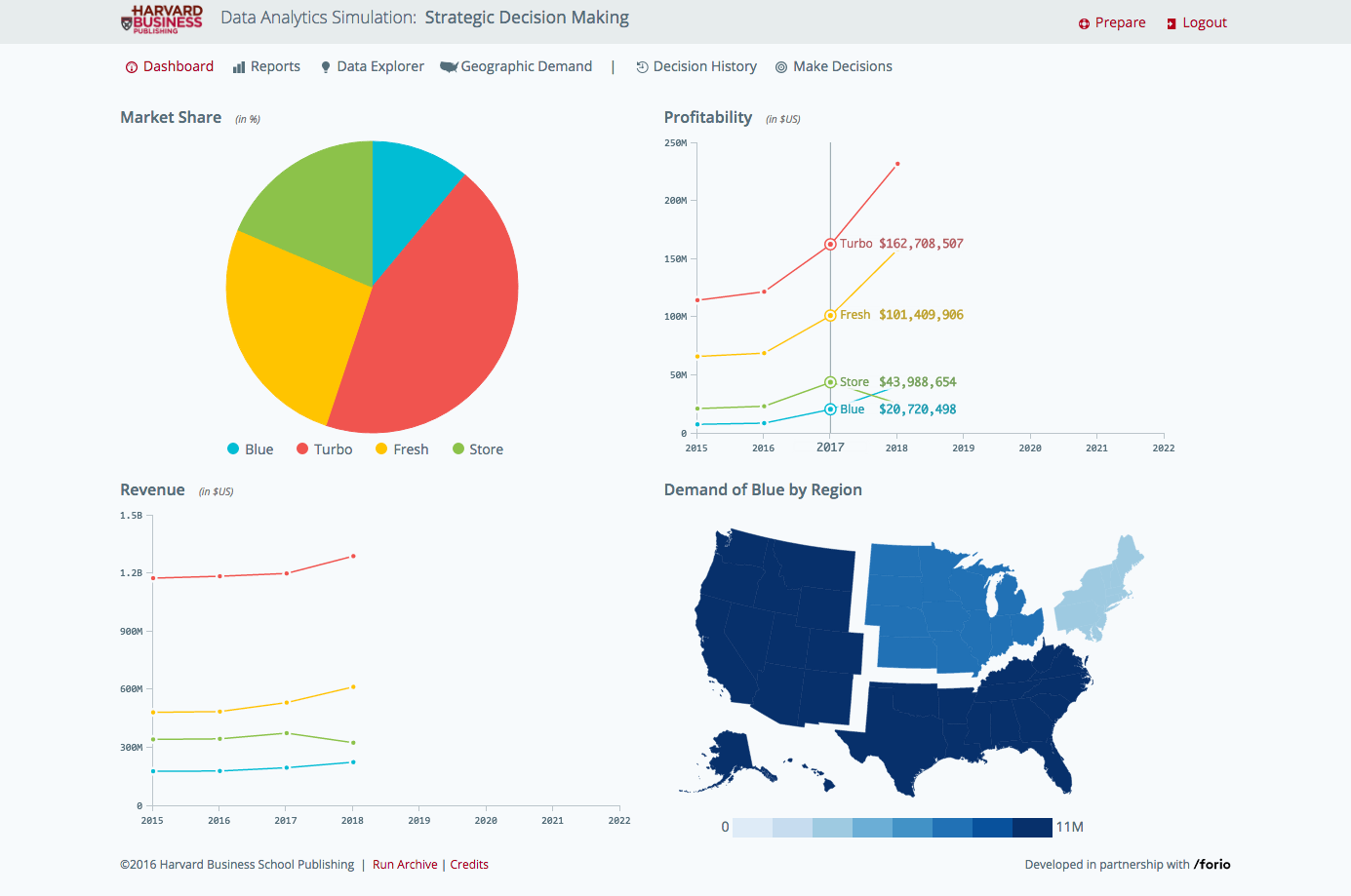
Created by Professor Tom Davenport, renowned thought leader on big data, this simulation teaches participants the power of analytics in decision-making. Acting as the brand manager for a laundry detergent, participants are tasked with turning around the brand's performance by using sophisticated analytic techniques to understand current issues. The process of StaR generation not only increases the stability of the receptor and its activity in a detergent environment, but because stabilisation is driven by the interaction of the receptor. Blue native gel analysis confirmed that ZDHHC6 can associate into higher order structures, which could be dimers and above, or associate with detergent resistant membrane domains. In contrast DHHC 7 and 20 appeared largely monomeric under the same solubilization conditions ( Figure 4D ). In this study, Active Queue Management Algorithms such as RED, ARED, SRED, REM, SBF, BLUE, RED, PURPLE, GREEN, CoDel used in mobile networks and the development of improved versions of these.
Abstract
Data integration is particularly difficult in neuroscience: we must organize vast amounts of data around only a few fragmentary functional hypotheses. It has often been noted that computer simulation, by providing explicit hypotheses for a particular system and bridging across different levels of organization, can provide an organizational focus which can be leveraged to form substantive hypotheses. Simulations lend meaning to data and can be updated and adapted as further data comes in. The use of simulation in this context suggests the need for simulator adjuncts to manage and evaluate data. We have developed a neural query system (NQS) within the NEURON simulator, providing a relational database system, a query function and basic data-mining tools. NQS is used within the simulation context to manage, verify and evaluate model parameterizations. More importantly, it is used for data-mining of simulation data and comparison to neurophysiology.

1 Introduction
Knowledge discovery and data-mining (KDD) is a process of seeking patterns among the masses of data that can be stored and organized in modern computer infrastructure. KDD arose in the commercial sector, where information was amassed for accounting and legal reasons over decades before being recognized as a valuable resource, figuratively a gold-mine of information. In the scientific realm, similar techniques have been developed and adapted over the past decade as massive information veins associated with the genomes of human, fly, mouse, and others have come on-line.

As suggested by its commercial history, data-mining grew out of databasing: data had been collected and now something was to be done with it. This history resulted in data-mining techniques arising apart from the underlying databasing approach. In this paradigm the database provides data for data-mining but is not itself altered by the data-mining endeavor (Fig. 1A). This limitation has been recently addressed by proposing the concept of inductive databases, which utilizes a two-way flow of information between database and data-mining tools.[1] The inductive database will include metadata calculated from the base data, which is then used as base data for further exploration.
Views of data-mining: A. Database centered; B. Simulator centered
Scientific data-mining differs in several ways from the more highly developed commercial applications.[2] Differences include a higher reliance on complex numerical manipulations and less clear-cut goals, requiring that data be analyzed and reanalyzed from different perspectives. In these respects, the scientific KDD process is more free-form than the commercial variety.
Neurobiologists are faced with the intellectual aim of understanding nervous systems, perhaps as complex a task as science faces. At the same time, modern techniques, including the genome projects, are making more and more information available, raising the question of what can best be done with it. In addition to the sociological perils common to cooperation in any scientific field, the emerging field of neuroinformatics must also confront unusual interoperability challenges due to the diverse types of data generated (see Note 1). These arise from different techniques, as well as from the many orders of magnitude in time and space covered by different investigations of even a single subsystem. [3]
Data-mining tools are typically chosen on an ad-hoc basis according to the task. These tools include various algorithmic constructions as well as traditional statistical techniques. Although some statistical procedures are used, the data-mining enterprise differs from statistics in that the data are multi-valued and do not generally fit a recognizable statistical distribution.[4] Thus statistical procedures that depend on the distribution, such as the Student’s t-test, often cannot be used. Those that are used are non-parametric. For example, mean and standard deviation lose meaning when working with a bimodal distribution.
Many data-mining tools in the commercial world are text-oriented or symbolic, providing clustering or classifications by meaning or symbols.[5] This symbol-oriented approach extends into science in the realm of genomics and proteomics, whose objects of interest are described by a circumscribed list of symbols representing nucleotides and amino-acids.[, ] In neuroscience, spatially oriented tools, some developed from geography, can be utilized in areas such as development,[] neuroanatomy,[, , 11] and imaging.[, ] Taxonomic thinking, involving ontologies and part-whole relations, require semantically-oriented tools.[] Other areas of neuroscience, such as electrophysiology, are primarily numerical. Numerical data-mining tools include numerical integration and differentiation, [, ] wavelet and spectroscopic analyses, [17] numerical classification methods such as principal component and independent component analysis[] and standard statistical methods. Spike trains, being a time-series of discrete events, require yet other approaches for analysis.[19]
On the algorithmic side, iterative or recursive search programs are used to cluster or associate data or to make decision trees. Another major class of algorithm involves machine learning algorithms such as simulated annealing, genetic algorithms and artificial neural networks (ANNs). The category of artificial neural networks has potential for confusion in the present context. Realistic neural simulation includes realistic neural networks which seek to replicate the actual physiology of a set of connected neurons. By contrast, ANNs are typically used as tools for finding regularities in data. Although the original inspiration for ANNs came in part from neuroscience, these networks are not primarily used as direct models of nervous systems.
We have developed database and data-mining facilities in a Neural Query System (NQS) implemented within the NEURON simulation program (see Note 2). We suggest that realistic simulation of neural models will be the ultimate excavator in the data-mining endeavor, providing causality in addition to correlation.[20] One use of NQS is to manage models: to analyze existing simulations and to assist in development of new models. However, the main use for this package will be analysis of large-volume experimental data with both retrospective and online analysis and correlation of simulations related to such data.
2 Materials
NQS compiles into the NEURON simulator as a NMOD module containing C-code for rapid processing. A hoc module is then loaded that makes use of these low-level procedures. The NQS package is available at http://senselab.med.yale.edu/senselab/SimToolDB/default.asp. A user manual is included with the package.
3 Methods
As noted above, traditional data flow is one-way from databases to data-mining tools (Fig. 1A). The loop closes as data-mining insights are used to develop hypotheses that suggest new experiments. In this traditional view, simulation would be considered as just another data-mining tool, to be called from the data-mining suite in order to assess correlations. However, realistic simulation differs from other tools in that it provides causal explanations rather than simple correlations. For this reason, the simulator is intercalated in the hypothesis return loop in Fig. 1B. In addition to testing hypotheses generated by mining experimental data, simulation also generates new hypotheses, either directly or via mining of simulation output.
NQS is a simulator-based and simulator-centric databasing system with added data-mining tools. It is not a full database management system since it is meant primarily as single-user system that does not have to handle problems of data-access control and data security. It also does not presently have the indexing and hashing capabilities of a full database management system, thought these may be added in the future. NQS provides some spreadsheet functionality. However, several features characterize it as a database rather than a spreadsheet system: the presence of a query language; the ability to handle many thousands of records; data structuring; handling of non-numeric data; capacity for relational organization across database tables.
Using the simulator to control embedded databasing and data-mining software facilitates the use of simulation as a focusing tool for knowledge discovery (Fig. 1B). Several considerations suggest such a central role for neural simulation. First, there is the practical consideration of data quantity. Neural simulation, as it becomes more and more realistic, must become a massive consumer of experimental data, requiring a reliable pipeline. In addition to being dependent on experiment, simulation is itself an experimental pursuit, differing in this respect from the closed-form analytic models of traditional physics.[21] As we experiment on these models, the simulator becomes a producer of vast quantities of simulation data which must also be managed and organized.

In Fig. 1B, the two-headed arrow between “Simulation” and “Database” denotes not only the flow of experimental and simulation data but also the management of simulation parameters. Simulation parameters can be stored in NQS databases, separated from experimental and simulation data. Neural simulations, particularly network simulations, are highly complex and can be difficult to organize. Once set-up, it can be hard to visualize the resulting system in order to verify that the system has been organized as planned. Storing parameters in a database is a valuable adjunct for checking and visualizing parameter sets and for storing and restoring them between simulations. Data-mining can then be used both for comparing parameters among sets of simulations and for relating changes in parameter sets to changes in model dynamics.
As compared to many data-mining tools, neural simulation tends to be very computationally intensive, particularly when large parameter searches are undertaken. (Machine learning and artificial neural network algorithms are also computationally intensive.) Providing databasing and data-mining tools within the simulator allows partial data analysis to be done at run-time. This permits simulation results to be immediately compared with salient experimental features discovered through data-mining. Simulations that are a poor match can then be aborted prematurely. Similarly, in a parameter learning context, using a terrain search or evolutionary algorithm, fitness can be determined on the fly by using a variety of measures calculated with a set of data-mining tools.
Many neuroscience applications generate spatial data that do not map directly onto the rectangular array used in relational database tables (see Note 3). Some data relations will be lost or obscured in remapping. NQS allows object storage in cells, permitting pointers and other indicators of non-rectangular relations. Completely non-rectangular data storage formats can be implemented as an adjunct to the database tables which then use pointers to indicate locations in this supplementary data geometry. For example, the dendritic tree is stored in NEURON as a tree structure with parent and children pointers. In the course of reducing the tree to rectangular form, a variety of rectangular representations can be used, which then use pointers to maintain correspondence with the original tree format. These object adjuncts are useful for drilling down into raw data when doing data-mining. However, it remains unclear how basic database functions such as sort and select can be extended to make use of such non-rectangular objects in a useful way.
3.1 NQS functionality
NQS is implemented as a series of low-level routines that operates on vectors that are maintained as the parallel columns of a table. NQS thereby provides access to a variety of vector (array) manipulation tools built into NEURON. These vector functions permit convolution, numerical differentiation and integration, and basic statistics. Additional data-mining tools have been added on by compiling C-language code that can be directly applied to the numerical vectors used as columns for a table. Vector-oriented C-language code is readily available from a variety of sources.[22, 23] Such code can be compiled into NEURON after adding brief linking headers.
NQS handles basic databasing functionality including: 1. creating tables; 2. inserting, deleting and altering tuples; 3. data queries. More sophisticated databasing functionality such as indexing and transaction protection are not yet implemented. Databasing commands in NQS provide 1. selection of specified data with both numerical and limited string criteria ; 2. numerical sorting; 3. printing of data-slices by column and row designators; 4. line, bar and scatter graphs; 5. import and export of data in columnar format; 6. symbolic spreadsheet functionality; 7. iterators over data subsets or over an entire table; 8. relational selections using criteria across related tables; 9. mapping of user specified functions onto particular columns.
Among basic databasing functions, querying is the most complex. A query language, although often regarded as a database component and thereby denigrated as a data-mining tool, is a critical aspect of data-mining. Structured Query Language (SQL), because of its commercial antecedents, is less numerically oriented than is desirable for scientific query. The NQS select command is designed to focus on numerical comparisons. Due to the importance of geometric information in neuroscience, inclusion of geometric criteria will be an additional feature that would be desirable in further development of NQS.
The NQS select() command is similar to the commands related to the WHERE and HAVING sub-functions of SQL’s SELECT. NQS syntax naturally differs from that of SQL as it must follow the syntax of NEURON’s object-oriented hoc language. An NQS database table is a template in hoc.
The NQS select() command takes any number of arguments in sets. Each set consists of a column name, a comparative operator such as ‘<’ or ‘’ and one or two arguments depending on the operator. Multiple criteria in a single select() statement are handled with an implicit AND. A flag can be set to use OR on the clauses. A command can also begin with “&&” or “||” to return the union or respectively intersection of the selected rows with previously selected rows (cf. SQL UNION, INTERSECT subcommands). Although the NQS select() does not replicate the agglutinative syntax of SQL SELECT, this functionality can be effected by serial application of NQS’s select(), sort() and stat() functions. Inner join between tables is implemented to permit formation of relational databases consisting of multiple tables.
3.2 Usage examples
We have been using NQS to explore system parameters, simulation results and the relation between parameters and simulation output. One seemingly trivial but important usage is to design, implement and verify the connectivity of a neuronal network. Although this task is fairly simple in a basic network which typically only has two classes of cells, one inhibitory and the other excitatory, the complexity increases enormously as multiple cell types are included, each with its own connectivity patterns with each other type. Individual tables can be developed for each postsynaptic cell type or region of network thus allowing networks to be built up incrementally. This has proved important for rapidly loading large models with tens of thousands of cells and tens of millions of synapses synapses.[] In this context a table columns identify presynaptic cell type and index as well as postsynaptic receptor type and dendritic location. Other implementation specific information, such as pointer to the actual connection object, can be included as additional columns.[]
The primary use of the NQS package is analysis of large datasets, derived from either simulation or physiology. Although NQS makes it easy to manage the large datasets, there remains much work to be done in providing adequate techniques for display and assessing of these high-dimensional systems. In Fig. 2, we are evaluating simulated field potential data from a population of 132,000 1200-cell network simulations generated by varying 13 parameters over 2-3 values apiece.[26] Although the simulated field was originally generated to permit ready comparison to physiology, it became apparent that the field provided a useful abstract of the data otherwise contained in a raster-plot or in multiple simulated intracellular records, being far easier to compare between simulations than either of the alternatives.
Graphical use of NQS. See text for details.
Given the simulated field potentials, we extract population spikes and classify by height, width and occurrence time in order to fill out NQS tables that allow summarized classification of each simulation result. In Fig. 2, we compare the attributes of two selected sub-populations representing pair-wise matched simulations differing by two alternative values (alleles) in one parameter (single neuron excitability). In Fig. 2 bottom, we graph a point for each simulation pair indicating the difference in duration of population spiking activity (x-axis) and the total number of population spikes (y-axis). In this case, the upper right quadrant represents the expected result: increased single-neuron excitability results in a greater number of spikes (positive y-axis) and a longer duration of activity (positive x-axis). The interesting observation here is that there are many simulation pairs in the other quadrants as well, representing reduction in duration or spike number with increased neuron excitability.
In addition to a textual select() command, NQS permits graphical selection by surrounding a rectangle or clicking on an individual point with mouse and cursor. In Fig. 2, we have clicked on a single pointer in the right upper quadrant which displays the associated simulation pair above (higher excitability in black, lower in red). This allows a rapid graphical search through hundreds of simulations selected according to their relationships across chosen attributes. Alternatively, we can map individual or paired simulations according to parameters rather than according to activity and thereby search in parameter-space rather than in collapsed state-space. Given the vast number of simulations in this case, it is also desirable to develop higher-order automated selection protocols to find simulations with particular attributes. For example, in Fig. 3, we select for simulations with late population spike activity. Here, each trace is graphed with its unique ascension number.
Simulations selected according to complex criteria: here the presence of late population spikes.
4 Notes
1. Computer ubiquity is driving biology, together with most other fields, into the realm of big science. Unlike the big science of the past where large groups had to be organized and mobilized at a particular site, modern computer communication will allow groups to form and re-form ad hoc around particular problems and particular data sets. This informatics revolution is particularly welcome in neurobiology, where the stretch between levels of organization is so great and the amount of potential data so overwhelming in depth, breadth and variety.
2. Simulation offers a good focal point for working with data. We have illustrated here a simulator-embedded system for database manipulation. However, it is clear that the huge databases that will be required in neurobiology will need dedicated full-bandwidth database software. It is envisioned that NQS will play an intermediate role, together with standard database software. NQS might be used independently for the type of parameter definition and verification illustrated in the examples above. When saving results from large numbers of simulations or comparing to results from many experiments, NQS would read and write to disk via other database software but would still be able to rapidly manipulate and assess data online to compare simulations and experimental results.
3. NQS stores data in a standard relational database format. This rectangular data structure has limitations compared to the dedicated internal data structures of a simulator such as NEURON. For example, NEURON stores a dendritic tree using pointers to create a tree-like format involving parent and child sections. However, major database systems, whether commercial or open-source, require data in a rectangular, column/row format. Therefore it is important to translate into these representations in order to communicate between simulators and databases. One advantage of finding a simple, easily stored representation for models would be the ability to readily move a given model from one simulator to another. As compared to the use of a model meta-language (an XML), a database would require a more rigid format but would provide more rapid access and exploration. []
Acknowledgments
The author wishes to thank Mike Hines and Ted Carnevale for continuing assistance with NEURON This research was sponsored by NIH (NS045612 and NS032187).
References
Data-driven Management Of Blue Detergent Simulation
Data Driven Management Of Blue Detergent Simulation Method